Hantaviruses: A Global
Disease Problem
Connie Schmaljohn* and Brian Hjelle†
*United States Army Medical Research Institute of Infectious
Diseases, Fort Detrick, Frederick, Maryland, USA; and
†University of New Mexico, Albuquerque, New Mexico, USA
| Hantaviruses are
carried by numerous rodent species throughout the world.
In 1993, a previously unknown group of hantaviruses
emerged in the United States as the cause of an acute
respiratory disease now termed hantavirus pulmonary
syndrome (HPS). Before then, hantaviruses were known as
the etiologic agents of hemorrhagic fever with renal
syndrome, a disease that occurs almost entirely in the
Eastern Hemisphere. Since the discovery of the
HPS-causing hantaviruses, intense investigation of the
ecology and epidemiology of hantaviruses has led to the
discovery of many other novel hantaviruses. Their
ubiquity and potential for causing severe human illness
make these viruses an important public health concern; we
reviewed the distribution, ecology, disease potential,
and genetic spectrum. |
The genus Hantavirus, family Bunyaviridae, comprises at
least 14 viruses, including those that cause hemorrhagic fever
with renal syndrome (HFRS) and hantavirus pulmonary syndrome
(HPS) (Table
1). Several tentative members of the genus are known, and
others will surely emerge as their natural ecology is further
explored. Hantaviruses are primarily rodent-borne, although other
animal species harboring hantaviruses have been reported. Unlike
all other viruses in the family, hantaviruses are not transmitted
by arthropod vectors but (most frequently) from inhalation of
virus-contaminated aerosols of rodent excreta (1).
Human-to-human transmission of hantaviruses has not been
documented, except as noted below.
| Table 1. Members of the genus
Hantavirus, family Bunyaviridae |
|
| Species |
Disease |
Principal Reservoir |
Distribution
of Virus |
Distribution of Reservoir |
|
| Hantaan (HTN) |
HFRSa |
Apodemus agrarius
(striped field mouse) |
China,
Russia, Korea |
C Europe south to Thrace, Caucasus,
& Tien Shan Mtns; Amur River through Korea to E
Xizang & E Yunnan, W Sichuan, Fujiau, &
Taiwan(China) |
Dobrava-Belgrade
(DOB) |
HFRS |
Apodemus flavicollis
(yellow-neck mouse) |
Balkans |
England & Wales, from NW Spain,
France, S Scandinavia through European Russia to Urals, S
Italy, the Balkans, Syria, Lebanon, & Israel |
| Seoul (SEO) |
HFRS |
Rattus norvegicus
(Norway rat) |
Worldwide |
Worldwide |
| Puumala (PUU) |
HFRS |
Clethrionomys
glareolus
(bank vole) |
Europe, Russia,
Scandinavia |
W Palearctic from France and Scandinavia
to Lake Baikai, south to N Spain, N Italy, Balkans,W
Turkey, N Kazakhstan, Altai & Sayan Mtns; Britain
& SW Ireland |
| Thailand (THAI) |
ndb |
Bandicota indica
(bandicoot rat) |
Thailand |
Sri Lanka, peninsular India to Nepal,
Burma, NE India, S China, Laos, Taiwan, Thailand, Vietnam
|
| Prospect Hill (PH) |
nd |
Microtus
pennsylvanicus
(meadow vole) |
U.S., Canada |
C Alaska to Labrador, including
Newfoundland & Prince Edward Island, Canada; Rocky
Mountains to N New Mexico, in Great Plains to N Kansas,
& in Appalachians to N Georgia, U.S. |
| Khabarovsk (KHB) |
nd |
Microtus fortis
(reed vole) |
Russia |
Transbaikalia Amur region; E China |
Thottapalayam
(TPM) |
nd |
Suncus murinus
(musk shrew) |
India |
Afghanistan, Pakistan, India, Sri Lanka,
Nepal, Bhutan, Burma, China, Taiwan, Japan, Indomalayan
Region |
| Tula (TUL) |
nd |
Microtus arvalis
(European common
vole) |
Europe |
Throughout Europe to Black Sea & NE
to Kirov region, Russia |
| Sin Nombre (SN) |
HPSc |
Peromyscus maniculatus
(deer mouse) |
U.S., Canada, |
Alaska Panhandle across N Mexico Canada,
south through most of continental U.S., excluding SE
& E seaboard, to southernmost Baja California Sur and
to NC Oaxaca, Mexico |
| New York (NY) |
HPS |
Peromyscus leucopus
(white-footed mouse) |
U.S. |
C and E U.S. to S Alberta & S
Ontario, Quebec & Nova Scotia, Canada; to N Durango
& along Caribbean coast to Isthmus of Tehuantepec
& Yucatan Peninsula, Mexico |
Black Creek Canal
(BCC) |
HPS |
Sigmodon hispidus
(cotton rat) |
U.S. |
SE U.S., from S Nebraska to C Virginia
south to SE Arizona & peninsular Florida; interior
& E Mexico through Middle America to C Panama; in
South Amer ica to N Colombia & N Venezuela |
El Moro Canyon
(ELMC)d |
nd |
Reithrodontomys
megalotis
(Western harvest mouse) |
U.S., Mexico |
British Columbia & SE Alberta,
Canada; W and NC U.S., S to N Baja California &
interior Mexico to central Oaxaca |
| Bayou (BAY)d |
HPS |
Oryzomys palustris
(rice rat) |
U.S. |
SE Kansas to E Texas, eastward to S New
Jersey & peninsular Florida |
|
| Probable species:e
|
|
| |
| Topografov (TOP) |
nd |
Lemmus sibiricus
(Siberian lemming) |
Siberia |
Palearctic, from White Sea, W Russia, to
Chukotski Peninsula, NE Siberia, & Kamchatka;
Nearctic, from W Alaska E to Baffin Island & Hudson
Bay, S Rocky Mtns to C B.C., Canada |
| Andes (AND)d |
HPS |
Oligoryzomys
longicaudatusf
(long-tailed pygmy
rice rat) |
Argentina |
NC to S Andes, approximately to 50 deg S
latitude, in Chile & Argentina |
| |
| To be namedd |
HPS |
Calomys laucha
vesper mouse |
Paraguay |
N Argentina & Uruguay, SE Bolivia, W
Paraguay, and WC Brazil |
| Isla Vista (ISLA)d |
nd |
Microtus californicus
(California vole) |
U.S. |
Pacific coast, from SW Oregon through
California, U.S., to N Baja California, Mexico |
Bloodland Lake
(BLL)d |
nd |
Microtus ochrogaster
(prairie vole) |
U.S. |
N & C Great Plains, EC Alberta to S
Manitoba, Canada, S to N Oklahoma & Arkansas, E to C
Tennessee & W West Virginia, U.S.; relic populations
elsewhere in U.S. & Mexico |
| Muleshoe (MUL)d |
nd |
Sigmodon hipidus
(cotton rat) |
U.S. |
See Black Creek Canal |
| Rio Segundo (RIOS)d |
nd |
Reithrodontomys
mexicanus
(Mexican harvest mouse) |
Costa Rica |
S Tamaulipas & WC Michoacan, Mexico,
S through Middle American highlands to W Panama; Andes of
W Colombia & N Ecuador |
| Rio Mamore (RIOM)d |
nd |
Oligoryzomys microtis
(small-eared pygmy
rice rat) |
Bolivia |
C Brazil south of Rios Solimoes- Amazon
& contiguous low lands of Peru, Bolivia, Paraguay,
& Argentina. |
|
aHFRS,
hemorrhagic fever with renal syndrome
bnd, none documented
cHPS, hantavirus pulmonary syndrome
d not yet isolated in cell culture
e viruses for which incomplete
characterization is available, but for which there is
clear evidence indicating that they are unique
f suspected host, but not confirmed
Adapted from (57,72)
and from (9,13,23,38,42,43,50-71)
|
The recognition of a previously unknown group of hantaviruses
as the cause of HPS in 1993 is an example of virus emergence due
to environmental factors favoring of the natural reservoir; a
larger reservoir increases opportunities for human infection. We
reviewed the global distribution of hantaviruses, their potential
to cause disease, and their relationships to each other and to
their rodent hosts.
History of HFRS and HPS
"Hemorrhagic fever with renal syndrome" denotes a
group of clinically similar illnesses that occur throughout the
Eurasian landmass and adjoining areas (2,3). HFRS includes
diseases previously known as Korean hemorrhagic fever, epidemic
hemorrhagic fever, and nephropathia epidemica (4). Although these
diseases were recognized in Asia perhaps for centuries, HFRS
first came to the attention of western physicians when
approximately 3,200 cases occurred from 1951 to 1954 among United
Nations forces in Korea (2,5). Other outbreaks of what is
believed to have been HFRS were reported in Russia in 1913 and
1932, among Japanese troops in Manchuria in 1932 (2,6), and in
Sweden in 1934 (7,8). In the early 1940s, a viral etiology for
HFRS was suggested by Russian and Japanese investigators who
injected persons with filtered urine or serum from patients with
naturally acquired disease (2). These studies also provided the
first clues to the natural reservoir of hantaviruses: the
Japanese investigators claimed to produce disease in humans by
injecting bacteria-free filtrates of tissues from Apodemus
agrarius or mites that fed on the Apodemus mice. Mite
transmission was never conclusively demonstrated by other
investigators, and it was not until 1978 that a rodent reservoir
for HFRS-causing viruses was confirmed by investigators who
demonstrated that patient sera reacted with antigen in lung
sections of wild-caught Apodemus agrarius and that the
virus could be passed from rodent to rodent (9). The successful
propagation of Hantaan (HTN) virus in cell culture in 1981
provided the first opportunity to study this pathogen
systematically (10). The history of HFRS has been explored
(2,11,12).
HPS was first described in 1993 when a cluster of cases of
adult fatal respiratory distress of unknown origin occurred in
the Four Corners region of the United States (New Mexico,
Arizona, Colorado, and Utah). The unexpected finding that sera
from patients reacted with hantaviral antigens was quickly
followed by the genetic identification of a novel hantavirus in
patients' tissues and in rodents trapped near patients' homes
(13-15).
Prevalence and Clinical Course
Approximately 150,000 to 200,000 cases of HFRS involving
hospitalization are reported each year throughout the world, with
more than half in China (16). Russia and Korea also report
hundreds to thousands of HFRS cases each year. Most remaining
cases (hundreds per year) are found in Japan, Finland, Sweden,
Bulgaria, Greece, Hungary, France, and the Balkan countries
formerly constituting Yugoslavia (16). Depending in part on which
hantavirus is responsible for the illness, HFRS can appear as a
mild, moderate, or severe disease (Table 2). Death rates range
from less than 0.1% for HFRS caused by Puumala (PUU) virus to
approximately 5% to 10% for HFRS caused by HTN virus (16). The
clinical course of severe HFRS involves five overlapping stages:
febrile, hypotensive, oliguric, diuretic, and convalescent; it is
not uncommon, however, for one or more of these stages to be
inapparent or absent. The onset of the disease is usually sudden
with intense headache, backache, fever, and chills. Hemorrhage,
if it occurs, is manifested during the febrile phase as a
flushing of the face or injection of the conjunctiva and mucous
membranes. A petechial rash may also appear, commonly on the
palate and axillary skin folds. Sudden and extreme albuminuria,
around day 4, is characteristic of severe HFRS. As the febrile
stage ends, hypotension can abruptly develop and last for hours
or days, during which nausea and vomiting are common. One-third
of deaths occur during this phase because of vascular leakage and
acute shock. Almost half of all deaths occur during the
subsequent (oliguric) phase because of hypervolemia. Patients who
survive and progress to the diuretic phase show improved renal
function but may still die of shock or pulmonary complications.
The final (convalescent) phase can last weeks to months before
recovery is complete (3,5,12).
| Table 2. Distinguishing clinical
characteristics for HFRS and HPS |
|
| Disease |
Pathogens |
Distinguishing Characteristics* |
|
HFRS (moderate-severe)
Death rate
1%-15% |
HTN, SEO,
DOB |
hemorrhage +++
azotemia/
proteinuria +++/++++
pulmonary capillary leak +/++
myositis +/+++
conjunctival injection ++/++++
eye pain/myopia ++/++++ |
HFRS (mild)
Death rate <1% |
PUU |
hemorrhage +
azotemia/
proteinuria +/++++
pulmonary capillary leak -/+
myositis +
conjunctival injection +
eye pain/myopia ++/++++ |
HPS (prototype)
Death rate >40% |
SN, NY |
hemorrhage +
zotemia/
proteinuria +
pulmonary capillary leak ++++
myositis -
conjunctival injection -/+
eye pain/myopia - |
| |
HPS (renal
variant)
Death rate>40% |
BAY, BCC,
Andes |
hemorrhage +
azotemia/
proteinuria ++/+++
pulmonary capillary leak +++/++++
myositis ++/++++
conjunctival injection -/++
eye pain/myopia - |
|
*Minimum/maximum occurrence
of the characteristic: - rarely reported;
+ infrequent or mild manifestation; ++, +++, ++++ more
frequent and severe manifestation. |
More than 250 cases of HPS have been reported throughout North
and South America. Although the disease has many features (e.g.,
a febrile prodrome, thrombocytopenia, and leukocytosis) in common
with HFRS (Table
2), in HPS capillary leakage is localized exclusively in the
lungs, rather than in the retroperitoneal space, and the kidneys
are largely unaffected. Most of the 174 cases of HPS in the
United States and Canada have been caused by Sin Nombre (SN)
virus. In HPS, death occurs from shock and cardiac complications,
even with adequate tissue oxygenation. Cases of HPS in the
southeastern United States, as well as many in South America, are
caused by a newly recognized clade (a group that shares a common
ancestor) of viruses that includes Bayou (BAY), Black Creek Canal
(BCC), and Andes viruses. As with HFRS, clinical differences can
be observed among patients with HPS caused by different
hantaviruses. For example, although HPS due to SN virus infection
can sometimes be associated with renal insufficiency after
prolonged hypoperfusion, renal impairment is only rarely observed
early in disease, and chemical evidence of skeletal muscle
inflammation (increased serum levels of the muscle enzyme
creatine kinase) is rare (17).
In contrast, both renal insufficiency and elevated creatine
kinase levels are observed at much higher frequency, although not
universally, with Andes, BAY, and BCC virus infections (18-20;
J. Davis, J. Cortes, and C. Barclay, pers. comm.). In an outbreak
of HPS recently described in Paraguay, a novel hantavirus,
carried by Calomys laucha, was identified as the etiologic
agent (21).
The relationship of this virus to other HPS-causing hantaviruses
remains to be established.
Ecology and Epidemiology
Hantavirus infection is apparently not deleterious to its
rodent reservoir host and is associated with a brisk antibody
response against the virion envelope and core proteins and
chronic, probably lifelong infection. In natural populations,
most infections occur through age-dependent horizontal route(s).
The highest antibody prevalence is observed in large (mature)
animals. A striking male predilection for hantavirus infection is
observed in some rodent species such as harvest mice and deer
mice, but not in urban rats (Rattus norvegicus) (22-24).
Horizontal transmission among cage-mates was experimentally
demonstrated (25),
but vertical transmission from dam to pup is negligible or absent
both in wild and experimental settings (22,24,25).
Outbreaks of hantaviral disease have been associated with
changes in rodent population densities, which may vary greatly
across time, both seasonally and from year to year. Cycles
respond to such extrinsic factors as interspecific competition,
climatic changes, and predation. Spring and summer outbreaks of
HFRS in agricultural settings in Asia and Europe are linked to
human contact with field rodents through the planting and
harvesting of crops (16,26).
PUU outbreaks in Scandinavia and the HPS outbreak in the Four
Corners region of the United States were associated with natural
rodent population increases, followed by invasion of buildings by
rodents (27,28).
The ecologic events that led to 1994 and 1996 outbreaks of Andes
virus-HPS in Patagonia, a region in southern South America, are
being investigated. Human interventions, such as the introduction
of Old World plant species (e.g., rosas mosquetas and Scottish
brougham) to Patagonia, with associated alteration in rodent
population dynamics, have been suggested as possible factors.
Recent fires and a mild winter in Argentina's Rio Negro and
Chubut Provinces may also have had a positive effect on the
carrier rodent, the colilargo, Oligoryzomys longicaudatus
(M. Christie and O. Pearson, pers. comm.).
Although the aerosol route of infection is undoubtedly the
most common means of transmission among rodents and to humans,
virus transmission by bite may occur among certain rodents (29)
and may also occasionally result in human infection (30)
(often inside a closed space, such as a rodent-infested grain
silo, garage, or outbuilding used for food storage).
Epidemiologic investigations have linked virus exposure to such
activities as heavy farm work, threshing, sleeping on the ground,
and military exercises. Indoor exposure was linked to invasion of
homes by field rodents during cold weather or to nesting of
rodents in or near dwellings (16,31).
Genetic sequencing of rodent- and patient-associated viruses has
been used to pinpoint the precise locations of human infections,
which has supported the role of indoor exposure in hantavirus
transmission (32,33).
Many hantavirus infections have occurred in persons of lower
socioeconomic status because poorer housing conditions and
agricultural activities favor closer contact between humans and
rodents. However, suburbanization, wilderness camping, and other
outdoor recreational activities have spread infection to persons
of middle and upper incomes.
Nosocomial transmission of hantaviruses has not been
documented until very recently (34)
and must be regarded as rare. However, viruses have been isolated
from blood and urine of HFRS patients, so exposure to bodily
fluids of infected persons could result in secondary
transmission. Only rarely have multiple North American HPS cases
been associated with particular households or buildings. During
recent outbreaks of HPS in South America, however, clustering of
cases in households and among personal contacts appeared to be
more common (M. Christie, pers. comm.). During a recent outbreak
of Andes-virus-associated HPS in Patagonia, a Buenos Aires
physician apparently contracted the infection after minimal
exposure to infected patient blood (34;
D.A. Pirola, pers. comm.). An adolescent patient in Buenos Aires
apparently contracted hantavirus infection from her parents, who
were infected in Patagonia. This unprecedented observation of
apparent person-to-person spread of a hantavirus clearly requires
laboratory confirmation, especially by careful comparative
analysis of the viral sequences (32,33).
Hantaviruses have also caused several laboratory-associated
outbreaks of HFRS. Laboratory-acquired infections were traced to
persistently infected rats obtained from breeders (35-37),
to wild-caught, naturally infected rodents (38-40),
or to experimentally infected rodents (39).
No illnesses due to laboratory infections have been reported
among workers using cell-culture adapted viruses, although
asymptomatic seroconversions have been documented (40).
Hantavirus Distribution and Disease-causing Potential
The worldwide distribution of rodents known to harbor
hantaviruses (Table
1) suggests great disease-causing potential. Each hantavirus
appears to have a single predominant natural reservoir. With rare
exception, the phylogenetic interrelationships among the viruses
and those of their predominant host show remarkable concordance (Figure;
41).
These observations suggest that hantaviruses do not adapt readily
to new hosts and that they are closely adapted for success in
their host, possibly because of thousands of years of
coexistence. As many as three hantaviruses can be found in a
particular geographic site, each circulating in its own rodent
reservoir, with no apparent evolutionary influence on one another
(42).
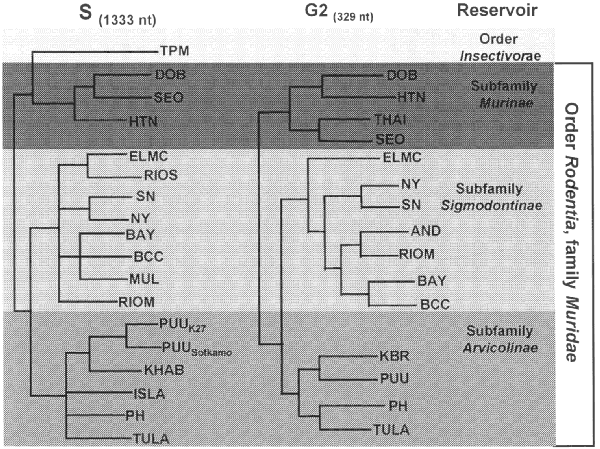
Figure. Phylogeny of hantaviruses and their
relationships to natural reservoirs. The trees were
constructed by comparing the complete coding regions of
the S segments of hantaviruses or of 330 nucleotides
corresponding to those of the M segment of Hantaan virus
(strain 76118) from nucleotides 1987 to 2315.
Abrreviations for viruses are as in Table 1. For each
analysis, a single most parsimonious tree was derived by
using PAUP 3.1.1 software. For the S segment tree,
boostrap values resulting from 100 replications were all
greater than 87% except for the branch leading to BCC
(78%) and the branch leading to DOB (52%). The next most
common placing of DOB was on a branch with HTN. |
All known hantaviruses, except Thotta-palayam (TPM) virus,
have been isolated or detected in murid rodents. Because only one
isolate of TPM virus was made from a shrew (Order Insectivora),
it is not clear if Suncus is the true primary reservoir or
an example of a "spillover" host, i.e., a secondary
host infected through contact with the primary host. Spillover is
common in sympatric murid rodents, including those identified as
the predominant carrier of another hantavirus; thus, the
opportunity for genetic exchange among hantaviruses is present in
nature. Spillover hosts are believed to have little or no impact
on hantaviral distribution or associated disease. However,
rodents other than the primary reservoirs can play an important
carrier role. For example, Microtus rossiaemeri-dionalis
may play a role in maintenance of Tula virus in some settings (43),
and Peromyscus leucopus and Peromyscus boylii can
be important reservoirs for SN virus in the western United States
(T. Yates and B. Hjelle, unpub. data). Apparent spillover may
also be the result of laboratory errors such as polymerase chain
reaction (PCR) contamination or misidentification of rodent
species. However, spillover is probably under-appreciated in many
studies that rely on reverse transcriptase PCR for identifying
specific viruses because many primer pairs may not detect an
unexpected spillover virus. In either case, because mistaken
identities and cell culture contaminations with other
hantaviruses have occasionally been reported, investigators
should verify unusual findings to prevent further confusion.
Antigenic and Genetic Diversity among Hantaviruses
Hantaviruses have been characterized by a combination of
antigenic and genetic methods. For viruses propagated in cell
culture, the plaque-reduction neutralization test is the most
sensitive serologic assay for differentiation (44,45);
nine hantaviruses have been defined by this test: HTN, Seoul
(SEO), PUU, Prospect Hill, Dobrava-Belgrade (DOB), Thailand, TPM,
SN, and BCC viruses (44-48).
Genetic relationships among hantaviruses are mirrored in their
antigenic properties. A direct correlation between genetic and
antigenic relationships is difficult; however, it can be assumed
that the plaque-reduction neutralization test measures
differences in the M segment gene products, i.e., the G1 and G2
envelope glycoproteins. Comparing the deduced G1 and G2 amino
acid sequences, therefore, may provide clues to the antigenic as
well as genetic diversity among hantaviruses.
Of characterized hantavirus isolates, SEO virus is the most
genetically homogeneous. Isolates of SEO virus, regardless of
their geographic origin, display M segment nucleotide and deduced
amino acid sequence homologies of approximately 95%, and 99%,
respectively (41,47).
A reported exception, the R22 isolate from China, had a slightly
lower homology; however, the original data suggest that an error
in the nucleotide sequence, resulting in a frame shift reading
error, may account for almost all of the additional changes. PUU
virus isolates vary the most, with M segment nucleotide and amino
acid sequence homologies of 83% and 94% observed between a
Finnish and Russian isolate. HTN virus also appears to be quite
stable in nature. Comparing the M segment sequences of prototype
HTN virus (from Apodemus) and those of two human isolates
obtained at a 6-year interval from HFRS patients in Korea
produced nucleotide and deduced amino acid sequence homologies of
94% and 97%, respectively (48).
For SN virus, comparing the complete M or S segment sequences of
three strains from California or New Mexico produced homologies
of 87% to 99%. Partial nucleotide sequence comparisons of the M
or S segments of SN viruses from adjacent counties in California,
detected in deer mice captured 19 years apart, were 97.5%
homologous (49).
Similarly, a retrospective analysis of archived tissue samples
collected in Mono County, California, in 1983 showed viruses with
partial M and S segment nucleotide sequence homologies of about
87% with SN from an 1993 HPS patient from New Mexico (50).
In all cases, the amino acid sequences encoded by these genes
differed between cognate proteins by much less than 5%. These
values are similar to those observed among strains of HTN virus.
Studies have just begun to appear in which the nature of
quasispecies in natural rodent hosts is defined (43,51).
Such investigations should provide more definitive data
concerning the genetic diversity among hantaviruses in nature.
Evolution of Hantaviruses
Phylogenetic trees derived by comparing complete or partial S
(Figure),
M, or L segment nucleotide sequences (41,52,53)
show two major lineages of hantaviruses, one leading to HTN, SEO,
Thailand, and DOB viruses, and one leading to PUU, Prospect Hill,
SN, and other New World hantaviruses. TPM virus, the first
hantavirus isolated in cell culture (54),
may be the most antigenically and genetically disparate member of
the genus; however, comparison of the complete nucleotide
sequence of the TPM S segment (A. Toney, B. Meyer, C. Schmaljohn,
unpub. data) suggests that TPM virus is more closely related to
HTN, SEO, and DOB viruses than to any of the other viruses in the
genus (Figure).
Nucleotide sequence homologies of the M and S segments of any two
hantaviruses have approximately the same degree of divergence
between each of the three segments, which suggests similar
evolutionary rates for these two gene segments. A slightly higher
homology among L segments sequenced to date perhaps indicates a
greater need for conservation of either RNA or protein functions
(47).
Point mutations appear to account for most of the genetic drift
among hantaviruses. Recombination has not been reported for
hantaviruses, although segment reassortment within a particular
species appears common (52,55).
The exchange of gene segments is suggested to be nonrandom, with
a higher propensity for M segment swapping, than for S or L (55).
Whether it contributes to the pathogenesis of hantaviruses is not
known, but reassortment certainly provides an avenue for more
rapid accumulation of changes than could occur by point mutation.
There is no evidence that reassortment can occur between
different species of hantaviruses; however, this has not been
studied systematically.
Murid rodents have probably harbored inapparent hantavirus
infections for thousands, perhaps millions of years. It is likely
that the genus Hantavirus evolved in the Old World and
that viruses were carried by rodents across the Bering land
bridge when they migrated during the Oligocene, and into South
America in the Pliocene (71).
Humans are incidental hosts, the victims of spillover infections
from the natural host rodents. One of the two major forms of
hantaviral diseases is endemic in each hemisphere. Both HFRS and
HPS can be divided into distinct clinical subtypes, and the viral
strain is a key determinant of the severity and nature of the
clinical abnormalities. Not covered in this review are clinical
studies of HFRS and HPS patients, which suggest that pathogenesis
may be immunologic and may be mediated by cytokine responses (72).
New outbreaks with novel hantavirus strains are still being
uncovered, especially in South America. However, the largest
clinical caseload and largest number of deaths occur in Asia and
Europe.
Dr. Schmaljohn is chief, Department of Molecular Virology,
USAMRID. Current research interests
include the development of molecular vaccines for hantaviruses,
filoviruses, and flaviviruses.
Dr. Hjelle has been active in studies of the molecular biology,
evolution, epidemiology, and clinical aspects of hantavirus
disease. His laboratory is a reference diagnostic center for
hantavirus infections of humans and animals and has recently
received funding to develop innovative vaccine strategies against
HPS and other emerging viral diseases.
Address for correspondence: Connie Schmaljohn, Virology
Division, USAMRID, Fort Detrick, Frederick, MD 21702-5011; fax:
301-619-2439; e-mail: cschmaljohn@detrick.army.mil
References
- Lee H, van der Groen G. Hemorrhagic fever with renal
syndrome. Prog Med Virol 1989;36:62-102.
- Gajdusek D. Virus hemorrhagic fevers. J Pediatr
1962;60:841-57.
- World Health Organization. Haemorrhagic fever with renal
syndrome: memorandum from a WHO meeting. Bull World
Health Organ 1983;61:269-75.
- Gajdusek DC, Goldfarb LG, Goldgaber D. Bibliography of
hemorrhagic fever with renal syndrome. Second Edition.
Bethesda (MD): National Institutes of Health; 1987 Pub
No. 88-3603.
- Smadel J. Epidemic hemorrhagic fever. Am J Public Health
1953;43:1327-30.
- Casals J, Henderson BE, Hoogstraakm G. A
review of Soviet viral hemorrhagic fevers. J Infect
Dis 1969;122:437-53.
- Zetterholm SG. Akuta nefriter simulerande akuta bukfall.
Svenska Lakartidningen 1934;31.
- Myhrman G. En njursjukdom med egenartad symptombild. Nord
Med Tidskr 1934;7:793-4.
- Lee H, Lee P, Johnson K. Isolation
of the etiologic agent of Korean hemorrhagic fever. J
Infect Dis 1978;137:298-308.
- French G, Foulke R, Brand O, Eddy G. Korean hemorrhagic
fever: propagation of the etiologic agent in a cell line
of human origin. Science 1981;211:1046-8.
- Traub R. Korean
hemorrhagic fever. J Infect Dis 1978;138:267-72.
- McKee K, LeDuc J, Peters C. Hantaviruses. In: Belshe RB,
Belshe RB, editors. Textbook of human virology. St Louis:
Mosby Year Book 1991:615-32.
- Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek
TG, Feldmann H, et al. Genetic
identification of a hantavirus associated with an
outbreak of acute respiratory illness. Science
1993;262:914-7.
- Ksiazek TG, Peters CJ, Rollin PE, Zaki S, Nichol S,
Spiropoulou C, et al. Identification
of a new North American hantavirus that causes acute
pulmonary insufficiency. Am J Trop Med Hyg
1995;52:117-23.
- Schmaljohn AL, Li D, Negley DL, Bressler DS, Turell MJ,
Korch GW, et al. Isolation
and initial characterization of a newfound hantavirus
from California. Virology 1995;206:963-72.
- Lee HW. Epidemiology and pathogenesis of hemorrhagic
fever with renal syndrome. In: Elliott RM, editor. The
Bunyaviridae. New York: Plenum Press, 1996:253-67.
- Duchin JS, Koster FT, Peters CJ, Simpson GL, Tempest B,
Zaki SR, et al. Hantavirus
pulmonary syndrome: a clinical description of 17 patients
with a newly recognized disease. N Engl J Med
1994;330:949-55.
- Khan AS, Spiropoulou CF, Morzunov S, Zaki SR, Kohn MA,
Nawas SR, et al. Fatal
illness associated with a new hantavirus in Louisiana.
J Med Virol 1995;46:281-6.
- Khan AS, Gaviria JM, Rollin PE, Hlady WG, Ksiazek TG,
Armstrong LR, et al. Hantavirus
pulmonary syndrome in Florida: association with the newly
identified Black Creek Canal virus. Am J Med
1996;100:46-8.
- Hjelle B, Goade D, Torrez-Martínez N, Lang-Williams M,
Kim J, Harris RL, et al. Hantavirus
pulmonary syndrome, renal insufficiency and myositis
associated with infection by Bayou hantavirus. Clin
Infect Dis 1996;23:495-500.
- Williams R, Bryan R, Mills H, Palma I, Vera I, Velasquez
F, et al. Hantavirus pulmonary syndrome in western
Paraguay. Am J Trop Med Hyg 1996;55 (suppl), Abstract 30.
- Mills J, Ksiazek T, Ellis B, Rollin P, Nichol S, Yates T,
et al. Patterns of association with host and habitat:
antibody reactivity with Sin Nombre virus in small
mammals in the major biotic communities of the
southwestern United States. Am J Trop Med Hyg. In press.
- Hjelle B, Anderson B, Torrez-Martínez N, Song W, Gannon
WF, L. YT. Prevalence
and geographic genetic variation of hantaviruses of New
World harvest mice (Reithrodontomys):
identification of a divergent genotype from a Costa Rican
Reithrodontomys mexicanus. Virology
1995;207:452-9.
- Childs J, Korch G, Glass G, LeDuc J, Shah K. Epizootiology
of hantavirus infections in Baltimore: isolation of a
virus from Norway rats and characteristics of infected
rat populations. Am J Epidemiol 1987;126:55-68.
- Lee H, Lee P, Baek L, Song C, Seong I. Intraspecific
transmission of Hantaan virus, etiologic agent of Korean
hemorrhagic fever, in the rodent Apodemus agrarius.
Am J Trop Med Hyg 1981;30:1106-12.
- Chen H-X, Qiu F-X, Dong B-J, Ji S-Z, Li Y-T, Wang Y, et
al. Epidemiologic studies on hemorrhagic fever with renal
syndrome in China. J Infect Dis 1986;154:394-8.
- Niklasson B, LeDuc J. Epidemiology of nephropathia
epidemica in Sweden. J Infect Dis 1987;155:269-76.
- Parmenter R, Vigil R. The hantavirus epidemic in the
southwest: an assessment of autumn rodent densities and
population demographics in central and northern New
Mexico. Atlanta (GA): 1993 Report to the Federal Centers
for Disease Control and Prevention.
- Glass G, Childs J, Korch G, LeDuc J. Association
of intraspecific wounding with hantaviral infection in
wild rats (Rattus norvegicus). Epidemiol
Infect 1988;101:459-72.
- Dournon E, Moriniere B, Matheron S, Girard P, Gonzalez J,
Hirsch F, et al. HFRS after a wild rodent bite in the
hautesavoie--and risk of exposure to Hantaan-like virus
in Paris laboratory. Lancet 1984;1:676-7.
- Armstrong L, Zaki S, Goldoft M, Todd R, Khan A, Khabbaz
R, et al. Hantavirus
pulmonary syndrome associated with entering or cleaning
rarely used, rodent-infested structures. J Infect Dis
1995;172:1166.
- Hjelle B, Torrez-Martínez N, Koster FT, Jay M, Ascher
MS, Brown T, et al. Epidemiologic
linkage of rodent and human hantavirus genomic sequences
in case investigations of hantavirus pulmonary syndrome.
J Infect Dis 1996;173:781-6.
- Jay M, Hjelle B, Davis R, Ascher M, Baylies HN, Reilly K,
et al. Occupational
exposure leading to hantavirus pulmonary syndrome in a
utility company employee. Clin Infect Dis
1996;22:841-4
- Enria D, Padula P, Segura EL, Pini N, Edelstein A, Riva
Posse C, et al. Hantavirus pulmonary syndrome in
Argentina: possibility of person to person transmission.
Medicina (B. Aires) 1996;56:709-11.
- Umenai T, Lee P, Toyoda T, Yoshinaga K, Lee H, Saito T,
et al. Korean
hemorrhagic fever in staff in an animal laboratory.
Lancet 1979;1:1314-6.
- Desmyter J, LeDuc J, Johnson K, Brasseur F, Deckers C,
Van Ypersele de Strihou C. Laboratory
rat associated outbreak of haemorrhagic fever with renal
syndrome due to Hantaan-like virus in Belgium. Lancet
1983;2:1445-8.
- Lloyd G, Bowen E, Jones N. HFRS
outbreak associated with laboratory rats in UK.
Lancet 1984;May:1775-6.
- Brummer-Korvenkontio M, Vaheri A, von Bonsdorff C-H,
Vuorimies J, Manni T, Penttinen K, et al. Nephropathia
epidemica: detection of antigen in bank voles and
serologic diagnosis of human infection. J Infect Dis
1980;141:131-4.
- Lee H, Johnson K. Laboratory-acquired
infections with hantaan virus, the etiologic agent of
Korean hemorrhagic fever. J Infect Dis
1982;146:645-51.
- Centers for Disease Control and Prevention. Laboratory
management of agents associated with hantavirus pulmonary
syndrome: interim biosafety guidelines. MMWR Morb
Mortal Wkly Rept 1994; 43:RR-7,1-2.
- Xiao SY, Leduc JW, Chu YK, Schmaljohn CS. Phylo-genetic
analyses of virus isolates in the genus Hantavirus,
family Bunyaviridae. Virology 1994;198:205-17.
- Rawlings J, Torrez-Martínez N, Neill S, Moore G, Hicks
B, Pichuantes S, et al. Cocirculation
of multiple hantaviruses in Texas, with characterization
of the S genome of a previously-undescribed virus of
cotton rats (Sigmodon hispidus). Am J Trop Med
Hyg 1996;55:672-9.
- Plyusnin A, Vapalahti O, Lankinen H, Lehvaslaiho H,
Apekina N, Myasnikov Y, et al. Tula
virus: a newly detected hantavirus carried by European
common voles. J Virol 1994;68:7833-9.
- Schmaljohn C, Hasty S, Dalrymple J, LeDuc J, Lee H, von
Bonsdorff C-H, et al. Antigenic
and genetic properties of viruses linked to hemorrhagic
fever with renal syndrome. Science 1985;227:1041-4.
- Chu Y-K, Rossi C, LeDuc J, Lee H, Schmaljohn C, Dalrymple
J. Serological
relationships among viruses in the Hantavirus genus,
family Bunyaviridae. Virology 1994;198:196-204.
- Chu Y-K, Jennings G, Schmaljohn A, Elgh F, Hjelle B, Lee
HW, et al. Cross-neutralization
of hantaviruses with immune sera from
experimentally-infected animals and from hemorrhagic
fever with renal syndrome and hantavirus pulmonary
syndrome patients. J Infect Dis 1995;172:1581-4.
- Schmaljohn CS. Molecular Biology of Hantaviruses. In:
Elliott RM, editor. The Bunyaviridae. New York: Plenum
Press, 1996:63-90.
- Schmaljohn C, Arikawa J, Hasty S, Rasmussen L, Lee WH,
Lee PW, Dalrymple J. Conservation
of antigenic properties and sequences encoding the
envelope proteins of prototype hantaan virus and two
virus isolates from Korean haemorrhagic fever patients.
J Gen Virol 1988;69:1949-55.
- Jay M, Ascher MS, Chomel BB, Madon M, Sesline D, Enge B,
et al. Sero-epidemiological studies of hantavirus
infection among wild rodents in California. Emerg Infect
Dis 1997;3.
- Nerurkar VR, Song JW, Song KJ, Nagle JW, Hjelle B,
Jenison S, et al. Genetic
evidence for a hantavirus enzootic in deer mice (Peromyscus
maniculatus) captured a decade before the recognition
of hantavirus pulmonary syndrome. Virology
1994;204:563-8.
- Rowe JE, St Jeor SC, Riolo J, Otteson EW, Monroe MC,
Henderson WW, et al. Coexistence
of several novel hantaviruses in rodents indigenous to
North America. Virology 1995;213:122-30.
- Li D, Schmaljohn A, Anderson K, Schmaljohn C. Complete
nucleotide sequences of the M and S segments of two
hantavirus isolates from California: evidence for
reassortment in nature among viruses related to
hantavirus pulmonary syndrome. Virology
1995;206:973-83.
- Spiropoulou CF, Morzunov S, Feldmann H, Sanchez A, Peters
CJ, Nichol ST. Genome
structure and variability of a virus causing hantavirus
pulmonary syndrome. Virology 1994;200:715-23.
- Carey D, Reuben R, Panicker K, Shope R, Myers R. Thottapalayam
virus: a presumptive arbovirus isolated from a shrew in
India. Indian J Med Res 1971;59:1758-60.
- Henderson WW, Monroe MC, St Jeor SC, Thayer WP, Rowe JE,
Peters CJ, et al. Naturally
occurring Sin Nombre virus genetic reassortants.
Virology 1995;214:602-10.
- Hjelle B, Jenison S, Torrez-Martínez N, Yamada T, Nolte
K, Zumwalt R, et al. A
novel hantavirus associated with an outbreak of fatal
respiratory disease in the southwestern United States:
evolutionary relation-ships to known hantaviruses. J
Virol 1994;68:592-6.
- Hjelle B, Jenison S, Goade D, Green W, Feddersen R, Scott
A. Hantaviruses:
clinical, microbiologic, and epidemiologic aspects.
Crit Rev Clin Lab Sci 1995;32:469-508.
- Avsic-Zupanc T, Xiao SY, Stojanovic R, Gligic A, van der
Groen G, LeDuc JW. Characterization
of Dobrava virus: a Hantavirus from Slovenia, Yugoslavia.
J Med Virol 1992;38:132-7.
- Lee H, Baek L, Johnson K. Isolation
of hantaan virus, the etiologic agent of Korean
hemorrhagic fever from wild urban rats. J Infect Dis
1982;146:638-44.
- Elwell M, Ward G, Tingpalapong M, LeDuc J. Serologic
evidence of hantaan-like virus in rodents and man in
Thailand. Southeast Asian J Trop Med Public Health
1985;16:349-54.
- Lee P, Amyx H, Yanagihara R, Gajdusek D, Goldgaber D,
Gibbs C Jr. Partial
characterization of Prospect Hill virus isolated from
meadow voles in the United States. J Infect Dis
1985;152:826-9.
- Hörling J, Chizhikov V, Lundkvist Ĺ, Jonsson M, Ivanov
L, Dekonenko A, et al. Khabarovsk
virus: a phylogenetically and serologically distinct
hantavirus isolated from Microtus fortis trapped
in far-east Russia. J Gen Virol 1996;77:687-94.
- Elliott LH, Ksiazek TG, Rollin PE, Spiropoulou CF,
Morzunov S, Monroe M, et al. Isolation
of the causative agent of hantavirus pulmonary syndrome.
Am J Trop Med Hyg 1994;51:102-8.
- Song J-W, Baek L-J, Gajdusek DC, Yanagihara R,
Gavrilovskaya I, Luft BJ, et al. Isolation
of pathogenic hantavirus from white footed mouse (Peromyscus
leucopus). Lancet 1994;344:1637.
- Ravkov EV, Rollin PE, Ksiazek TG, Peters CJ, Nichol ST. Genetic
and serologic analysis of Black Creek Canal virus and its
association with human disease and Sigmodon hispidus
infection. Virology 1995;210:482-9.
- Torrez-Martínez N, Song W, Hjelle B. Nucleotide
sequence analysis of the M genomic segment of El Moro
Canyon hantavirus: antigenic distinction from Four
Corners hantavirus. Virology 1995;211:336-8.
- Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA,
Rollin PE, Ksiazek TG, et al. A
newly recognized virus associated with a fatal case of
hantavirus pulmonary syndrome in Louisiana. J Virol
1995;69:1980-3.
- Plyusnin A, Vapalahti O, Lundkvist A, Henttonen H, Vaheri
A. Newly
recognised hantavirus in Siberian lemmings [letter].
Lancet 1996;347:1835.
- López N, Padula P, Rossi C, Lázaro ME,
Franze-Fernández MT. Genetic
identification of a new hantavirus causing severe
pulmonary syndrome in Argentina. Virology
1996;220:223-6.
- Song W, Torrez-Martínez N, Irwin W, Harrison J, Davis R,
Ascher M, et al. Isla
Vista virus: a genetically novel hantavirus of the
California vole Microtus californicus. J Gen
Virol 1995;76:3195-9.
- Hjelle B, Torrez-Martínez N, Koster FT. Hantavirus
pulmonary syndrome-related virus from Bolivia. Lancet
1996;347:57.
- Mertz G, Hjelle B, Bryan R. Hantavirus infection. In:
Fauci A, Schrier R, editors. Advances in internal
medicine. Chicago: Mosby Year Book Inc., 1996:373-425.

Emerging Infectious Diseases
National Center for Infectious Diseases
Centers for Disease Control and Prevention
Atlanta, GA
URL: http://www.cdc.gov/ncidod/EID/vol3no2/schmaljo.htm
Updated: 97/05/14

